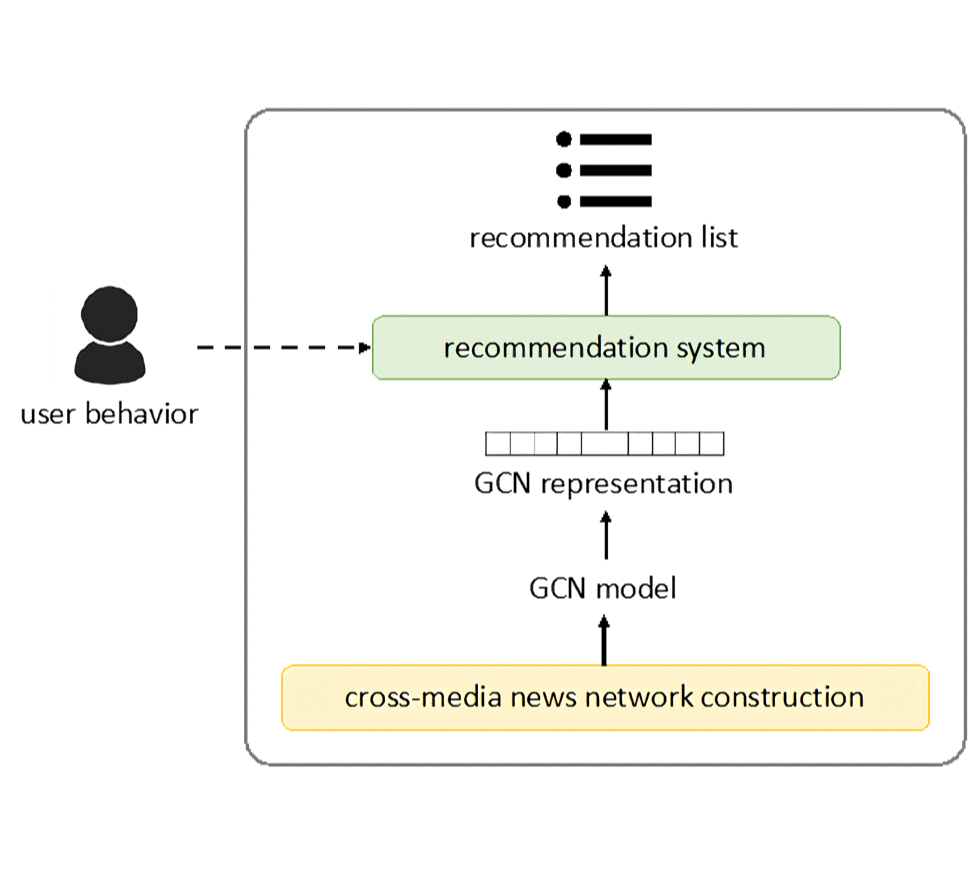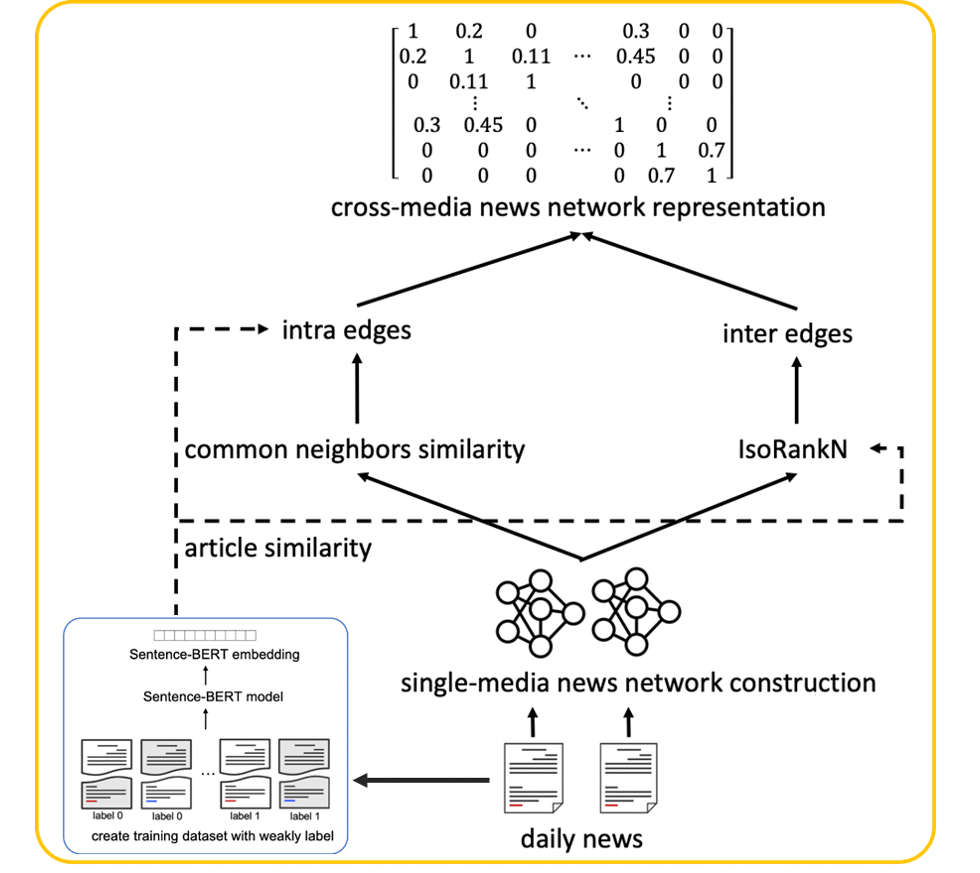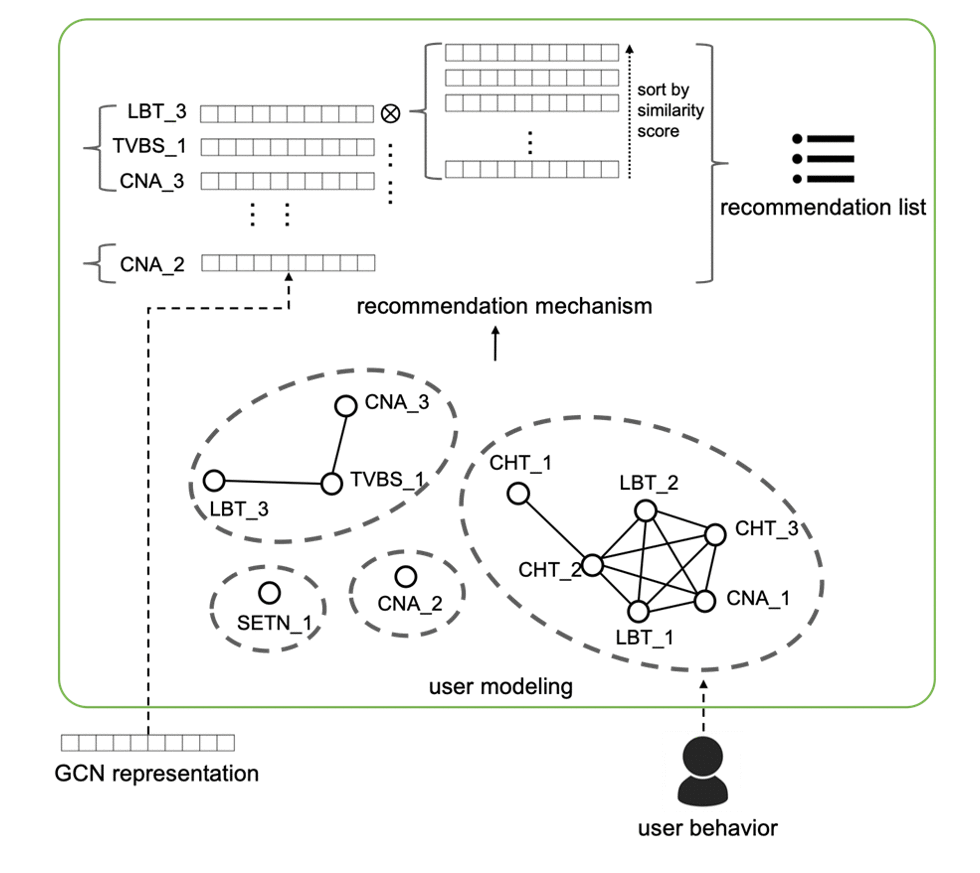
 Nowadays online recommendation systems aim at offering a wide range of up-to-date information and providing personalized recommendations since people usually read news shortly. However, existing studies either recommend news from the same media platform or just bring multiple sources of information together without considering readers’ preferences. Sometimes the former may lead to the media bias problem or even the fake news spread.
Nowadays online recommendation systems aim at offering a wide range of up-to-date information and providing personalized recommendations since people usually read news shortly. However, existing studies either recommend news from the same media platform or just bring multiple sources of information together without considering readers’ preferences. Sometimes the former may lead to the media bias problem or even the fake news spread.
Based on our previous network-alignment approach, we propose a novel cross-media news recommendation system, which can provide articles with diverse perspectives from different news media and preserve both document and topological information based on the inter- and intra-connections between news articles. Moreover, according to user browsing history, the recommendations made by the system can not only help readers understand their personalized interested news from a broader overview but mitigate the media bias problem. Empirical experiments with real users demonstrate the effectiveness of our system, and reveals the personalization and diversity of the recommendations [1].
Moreover, network alignment can be extended to assist weakly supervised learning in multi-aspect review comment analysis. Notice that nowadays online users’ feedback usually comprises only a single comment-level score and a review comment on mainstream platforms, such as Yelp and Google Maps. Reading through numerous review comments to figure out the important aspects of a target item (e.g., food quality of a restaurant) is definitely time-consuming. Moreover, some missing aspect information is entangled in the text commentaries and hardly accessible. Apparently, there is a huge gap between available review information and crucial aspects for users’ preferences, while aggregating the aspect information by text analysis could become the key to success. There has been, however, a dearth amount of research on weakly supervised models for this topic. In this study, we thus proposed a weakly supervised framework called Seq2CASE (Sequence to Commentary Aspect Score Estimation) to estimate the vital aspect scores from the limited mainstream review information. The aspect score estimation from only the weakly supervised signals is close to the actual aspect scoring; precisely, the average MAE (Mean Absolute Error) is less than 0.4 for a 5-point grading scale. The performance of the weakly supervised Seq2CASE framework is comparable to or even better than the state-of-the-art supervised approaches in recommendation tasks. We further demonstrate the capability of analyzing users’ behavior by exploiting only Seq2CASE regardless of users’ background. We expect this work to be a stepping stone that can inspire more studies working on this important but relatively underexploited research topic [2].
Smart restaurant:
This is a real implementation of Seq2CASE for the review comment analysis of restaurants on Google Map. The preliminary version considers around 150,000 restaurants with 14 million text reviews in Taiwan as well as 15,000 restaurants with 3.5 million text reviews in New York City.
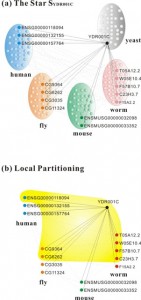 We also studied a fundamental problem in systems biology, i.e., to understand the cell as a system of interacting components and especially, almost every biological process is mediated by a network of molecular interactions. In particular, there has been a considerable amount of research devoted to the discovery and exploration of interactions between proteins in the last decade. Since many cellular activities are a result of protein interactions, proteins often interact with other proteins to perform their functions, and form a complex biological system, i.e., a protein-protein interaction (PPI) network. This powerful way of representing and analyzing the vast corpus of PPI data describes the interaction relationship among proteins in a cell. Furthermore, knowledge about the topology of a PPI network in one organism can yield insights about not only the networks of similar organisms, but also the function of their components. Hence comparison between protein interaction networks is becoming central to systems biology.
We also studied a fundamental problem in systems biology, i.e., to understand the cell as a system of interacting components and especially, almost every biological process is mediated by a network of molecular interactions. In particular, there has been a considerable amount of research devoted to the discovery and exploration of interactions between proteins in the last decade. Since many cellular activities are a result of protein interactions, proteins often interact with other proteins to perform their functions, and form a complex biological system, i.e., a protein-protein interaction (PPI) network. This powerful way of representing and analyzing the vast corpus of PPI data describes the interaction relationship among proteins in a cell. Furthermore, knowledge about the topology of a PPI network in one organism can yield insights about not only the networks of similar organisms, but also the function of their components. Hence comparison between protein interaction networks is becoming central to systems biology.
With the increasing availability of large PPI networks, comparative analysis of PPI networks across species has been proving to be a valuable method to study biology at a systems level. Protein interaction network alignment enables us to identify conserved functional components across species and perform high-quality ortholog prediction, i.e., identifying genes in different species derived from the same ancestral region. However, the computational complexity of alignment of multiple networks grows exponentially in the number of species. Besides, the genomes corresponding to the various networks being aligned may vary widely in size. We aim for the better global alignment algorithm of multiple PPI networks which will enable more accurate identification of functional orthologs across species more efficiently. We have collaborated with the MIT team and developed global alignment algorithms for performing comparative analysis of multiple biological networks, and especially our graph-theoretic spectral clustering technique, IsoRankN which is analogous to PageRank used by Google to rank web pages, outperforms all the existing algorithms so far. The related results have appeared as oral papers in the top conferences ISMB’09 [8] and PSB’10 [7], and one of the papers has been invited to be published in Bioinformatics [8].
In addition, the use of PPI networks in computing orthologs can produce biologically-appropriate mappings that better conserve protein function across species. It also facilitates annotation transfer from well-studied species to others. This practical use has also motivated a significant amount of work in the identification of orthologs. We consider the ortholog detection problem which is to identify gene correspondences across species that maximize functional similarity. Our goal is to build a publicly available network-based ortholog database that focuses on groupings of functionally related proteins. We have also collected the clusters computed using IsoRankN and build the first functionally ortholog (network-based) database, IsoBase (also see prototype). The related result has been accepted by the top journal Nucleic Acids Research [6].
We extend IsoRankN to explore the phylogenetic relationships between microorganisms through global alignment of multiple metabolic networks [5]. The resulting trees reflect the living style of organisms as well as classical taxa. For phylogenetically closely related organisms, the classification results are consistent with specific metabolic characteristics. Moreover, our exhaustive analysis of microbial metabolic pathways reveals differences in metabolic features between phylogenetically closely related organisms. We then proposed a new aligner, PISwap, for optimizing global pairwise alignments of protein interaction networks based on a local optimization heuristic [4]. PISwap can begin with different types of network alignment approaches and then iteratively adjust the initial alignments by incorporating network topology information, trading it off for sequence information. In practice, our algorithm efficiently refines other well-studied alignment techniques with almost no additional time cost. We also show the robustness of the algorithm to noise in protein interaction data.
Recently, we designed a new algorithm to automatically identify protein complexes [3], which is one of the most challenging tasks in systems biology. The algorithm integrates functional orthology information across multiple species to expand the search space of protein complex detections. As part of our approach, we also defined a new edge clustering coefficient to assign weight to interaction edges in PPI networks so that protein complexes can be identified more accurately. The edge clustering coefficient is based on the intuition that there is functional information captured in the common neighbors of the common neighbors as well. Our results showed that the algorithm outperforms well-known protein complex identification tools in a balance between precision and recall on three eukaryotic species: human, yeast, and fly. As a result of multiple network alignments of the species, the proposed approach can tolerate the edge loss of PPI networks and even discover sparse protein complexes which have traditionally been a challenge to predict.
1. Ting-Yu Kuo, Yi-Ping Phoebe Chen and Chung-Shou Liao. (2022) Network Alignment with Representation Learning for Personalized Cross-media News Recommendation System, submitted.
2. Chien-Tse Cheng, Yu-Hsun Lin and Chung-Shou Liao. (2022) Seq2CASE: Weakly Supervised Sequence to Commentary Aspect Score Estimation for Recommendation, submitted.
3. Cheng-Yu Ma, Yi-Ping Phoebe Chen, Bonnie Berger, Chung-Shou Liao. Identification of Protein Complexes by Integrating Multiple Alignment of Protein Interaction Networks, Bioinformatics, Vol. 33(11), (2017), pp. 1681-1688. (DOI: 10.1093/bioinformatics/btx043)
4. Leonid Chindelevitch, Cheng-Yu Ma, Chung-Shou Liao, and Bonnie Berger. Optimizing a Global Alignment of Protein Interaction Networks, Bioinformatics, Vol. 29(21), (2013) pp. 2765-2773.
5. Cheng-Yu Ma, Shu-Hsi Lin, Chi-Ching Lee, Chuan Yi Tang, Bonnie Berger and Chung-Shou Liao. Reconstruction of phyletic trees by global alignment of multiple metabolic networks, BMC Bioinformatics, Vol. 14 (S2) : S12, (2013) pp.1-9.
6. Daniel Park, Rohit Singh, Michael Baym, Chung-Shou Liao, and Bonnie Berger. IsoBase: a database of functionally related proteins across PPI networks, Nucleic Acids Research, Vol. 39 (2011) D295-D300.
7. Leonid Chindelevitch, Chung-Shou Liao, and Bonnie Berger. Local optimization for global alignment of protein interaction networks, Pacific Symposium on Biocomputing (PSB’10), Hawaii, U.S.A., 15, pp.123-132.
8. Chung-Shou Liao, Kanghao Lu, Michael Baym, Rohit Singh, and Bonnie Berger. IsoRankN: Spectral methods for global alignment of multiple protein networks, in Proceedings of the 17th International Conference on Intelligent Systems for Molecular Biology (ISMB’09), Stockholm, Sweden (The full version is invited to be published in Bioinformatics, Vol 25 No. 12 (2009) pp. i253-i258).
